NEUROPHOTONICS
Traditional in vivo neuroimaging is performed by magnetic resonance imaging (MRI) both in clinical and research settings. It is recognized as a highly useful tool in the study of the human brain, structurally, functionally and longitudinally. However, in vivo imaging restricts the resolution achievable which is coupled to the imaging time and is around 1 mm. The typical dimension we are looking for in the brain range from nanometers for the synapse, micrometer for the fibers, tens of micrometers for the neurons, hundreds of micrometers for cortical layers, millimeters for cortical depth and centimeter for cortical regions, not counting subcortical regions. Research has turned to ex vivo MRI imaging which can now reach down to 100 µm in resolution but some of the smaller scales are still missing. The gold standard to study the cytoarchitecture (cellular organization) and myeloarchitecture (fiber organization) of the human brain is histology, a century old technique, which yields high resolution and specificity but introduces high distortions which renders three-dimensional studies challenging.
In 2011, I was hired between two laboratories of the Athinoula A. Martinos Center for Biomedical Imaging, the Laboratory for Computational Neuromaging, specialized in MRI analysis, and the Optics Group, to use optical microscopy and image the post-mortem human brains for the first time. Optical imaging yields high resolution, down to the micrometer, allowing us theoretically to see individual neurons and myelinated fibers. Contrary to histology that requires the tissue to be sectioned prior to staining (the cause of the distortions), optical imaging is performed on the tissue block which greatly minimizes the deformations of the tissues and therefore allows the 3D reconstruction of the tissue, while preserving its spatial organization. Optical Coherence Tomography / Microscopy (OCT/OCM) has been developed in 1991 by Fujimoto et al. for the ophthalmology field but have been widely adopted since for in vivo rodent brain imaging.
- Ex vivo Human brain imaging validation
OCT relies on intrinsic optical properties of the tissue components, which in the brain mainly the myelinated fibers, neurons, vessels, glia, astrocytes and extracellular matrices. The cerebral cortex is divided into Brodmann areas (BA), which are closely correlated with the diverse cortical functions. The accurate segmentation of the human brain regions is paramount to understand the relationship between structure, function and connectivity. Those regions are defined based on their cytoarchitecture: the cortex is divided into six layers, whose thickness, density and cell type change with the regions. Moreover, the fiber input and output at each layer may also differ. Therefore, each cortical layer within one region has a unique composition, hence unique optical properties. Each region has its own set of layers, though some regions vary only slightly, while other are very unique. The gold standard to study the cytoarchitecture and define the boundary between regions is histology and more precisely Nissl staining which highlight healthy neurons. We have imaged various brain regions and identify these cortical regions. We have then compared these regions with the histological slices obtained on the same samples and showed high agreement while demonstrating the low distortions of OCT technique compared to histology.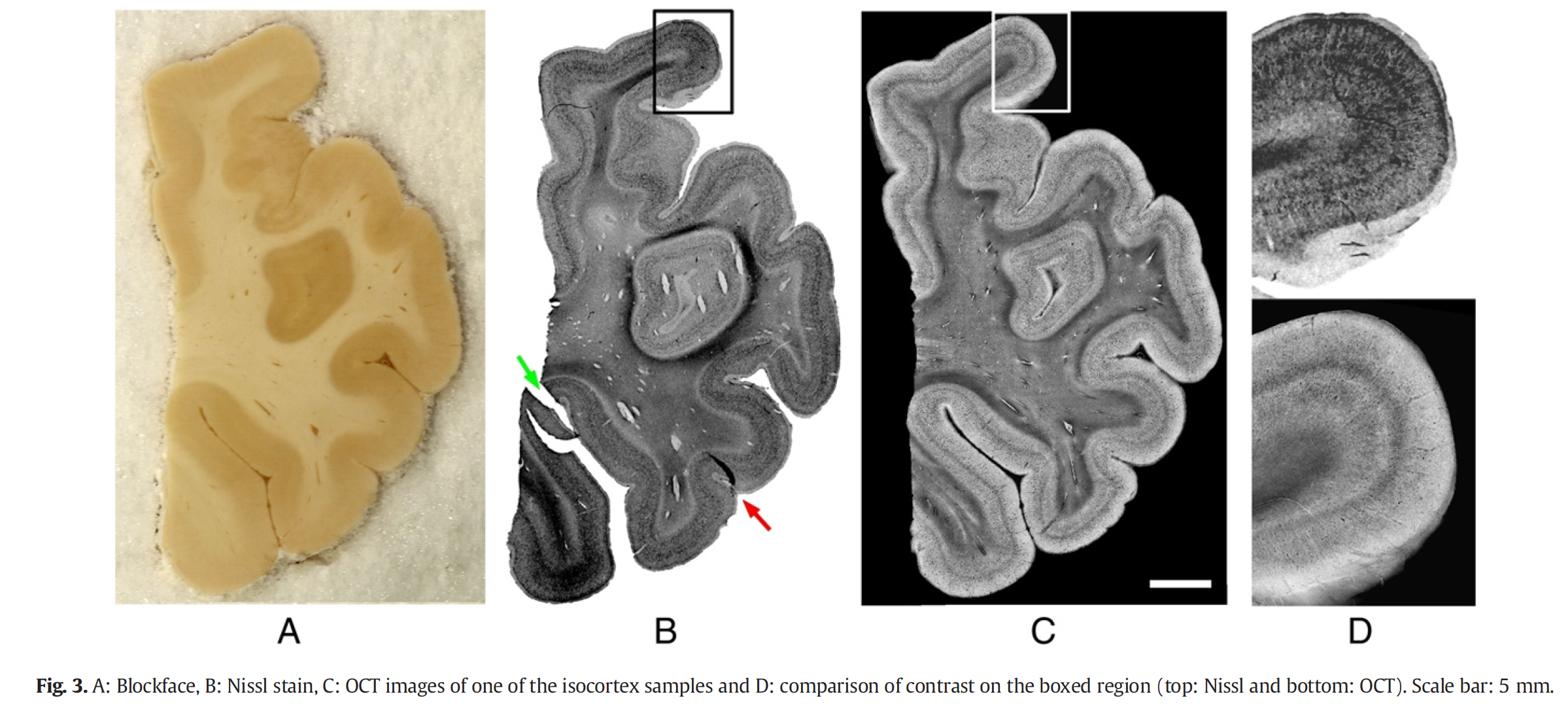
We have also pushed the comparison to histology at the individual neuron level using optical coherence microscopy (1 µm resolution) in the superficial layers of entorhinal cortex, a region targeted in Azheimer’s disease.We showed that OCM can reliably visualized the individual neurons. We have extended this validation to other regions of the brains. Other than the cytoarchitecture, we are greatly interested in imaging the myeloarchitecture of the brain using polarization coherence tomography, which relies on the birefringent property of the myelin sheath surrounding the fibers.
- Technical Advances
Beside validating OCT/OCM as a new imaging technique for neurosciences, we work on improving the technique both on the signal processing side and the acquisition side.
- Signal processing
As any interferometric technique, OCT/OCM suffers from speckle noise which can affect our ability to discern and, in the end, automatically segment the features of interest in our data such as the layers of the cortex or the neurons for example. We work on decreasing the noise and improving the signal to noise ratio in our imaging so we can develop algorithm that would automatically label OCT dataset using deep learning. Finally, OCT is an intensity-based method. We are working on quantifying the intrinsic optical properties of the brain tissue, mainly the scattering based on the measured depth intensity. The optical properties are related more specifically to the cell density, their size and shape, as well as the myelinated fibers size and density. This improvement is critical to elevate OCT to a quantification tool as well as an imaging tool.
- Large sample imaging
As any microscopy technique, the field of view is limited to a few millimeters at best in en-face area and to a few tens or hundreds of microns in depth. However, to image the human brain, or at least a region large enough to encapsulate a boundary between regions for example, a few regions, or an entire subcortical region, the sample need to be translated in the 3 directions and the superficial layer, previously imaged, has to be removed. With a collaboration with Tissue Vision Inc., we integrated 3-axis motorized stages and a vibratome to our step-up, as well as a customized software that automate the whole process: translation of the sample, OCT acquisition and sectioning. Moreover, since then, we have completely redeveloped the OCT acquisition software, to be fully customizable. We can now preprocess the data on the fly and most importantly reduce it size, which given the resolution of our resolution can be hard to managed.
- Multimodal integration: MRI, histology
While structural brain mapping has improved with high resolution ex vivo MRI, the resolution and contrast of MRI has limits that constrain our ability to visualize cytoarchitectural features in association cortices, even ex vivo. Refined localization of brain areas is critical for application to diseases such as autism, schizophrenia and Alzheimer’s disease, as well as functional MRI studies. Diffusion-weighted MRI (DW-MRI) allows us to probe the microstructure of the white matter (WM) by estimating the preferential directions of the diffusion of water molecules at every voxel. Although DW-MRI is now widely used to study WM integrity in health and disease, its validation has been challenging due to the absence of ground truth regarding the true connectivity of the brain. The ground truth for brain structural is the histology and it is also our main source neuropathological markers of diseases, such as cortical thinning, neural death, amyloid plaques for example. However, the high degree of non-linear distortions between histology slices and between histology and MRI render the interpretation of the correlation difficult. In our lab, we are using OCT/OCM as an intermediate tool. OCT can visualize the cortical layers and boundaries and can therefore be used to segment the brain. Due to its low deformation, we can register it, using a rigid transformation only, to any MRI modality to refine brain segmentation algorithm. PSOCT is used to directly validate DW-MRI algorithm and to disentangle the challenging regions of the brain where the fibers bundle either cross or come very close within one voxel. Finally, volumetric OCT imaging is utilized as an intermediate step between MRI and histology. Each slice corresponds to a known OCT slice acquisition which reduce the 3D non-linear registration problem to a 2D non-linear problem. OCT/OCM is a complementary technique to the existing neruoimaging techniques.
- Applications
Finally, I have collaborating with researchers from MGH and outside, on a wide range of applications, from tumors, to traumatic brain injuries, to neurodegenerative diseases.
- Infant brainstem and SIDS
Sudden infant death syndrome is the leading cause of postnatal infant mortality in industrialized nations. The arousal deficit hypothesis is the leading hypothesis and posits the cause of death as the failure in arousal in response to a life-threatening, sleep related stressor. The arousal ascending network and its pathways originate in the brainstem. Its nuclei and tracts are not visible by in vivo MRI and even high-resolution ex vivo MRI. Moreover, the lack of myelination in infants weakens the contrast of MRI. These limitations have greatly inhibited our ability to understand brainstem related diseases. Histology is used to localize the brainstem structures by staining neurons and/or fibers. However, this technique is labor intensive and suffers for irremediable distortions, rendering 3D reconstruction challenging and its precise registration to the corresponding ex vivo MRI for further analysis next to impossible. By using optical coherence tomography coupled with a vibratome, the entire infant brainstem can be imaged, without the need for dye or stain, at 2.9μm resolution prior to any sectioning. In OCT, distortions are greatly reduced, making 3D reconstruction and registration to ex vivo MRI data performed with a linear transformation straightforward. Even with the lack of myelination in the infant brain, OCT volume shows a great contrast, which allows the segmentation of the numerous nuclei and tracts of the brainstem. The creation of age-specific infant brainstem atlases will facilitate inference in ex vivo MRI and in in vivo MRI of connectivity emerging from the brainstem.
